Lab Alumni
- Nico Tenorio (Published!) | 2023-2024
- Victoria Colling (Published!) | 2022-2024
- Keaton Silver (Published!) | 2022-2024
- Audi Fineran (Published!) | 2022-2023
- Aidan Quinn | 2022-2023
- Dylan Poch: PhD student, Yale University (Published!) | 2019-2022
- Marissa Allen: PhD student, Marquette University | 2020-2022
- Mary Quansah (Published!) | 2021-2022
- Chris Chamblee: PhD student, Texas A&M | 2020-2022
- Andrew Smith: MD student, Kansas University (Published!) | 2019-2021
- Dr. Tyler Sodia: Particle Scientist, VitriVax; PhD, Colorado School of Mines (Published!) | 2018-2020
- Lindsey Armstrong: Lab technician, Creighton University | 2018-2020
- Austin Haider: PhD student, Denver University | 2018-2020
- Marcos Maldonado: (Published!) | 2016-2019
- Anika James: PhD student, Kansas University | 2018-2019
- Marlea Kudlauskas: Cardiac ICU Nurse | 2018-2018
- Anna Nguyen: PhD student, University of California Santa Barbara (Published!) | 2017-2018
- Lisa Fetter: PhD student, University of California Santa Barbara (Published!) | 2014-2018
- Derek Clark: Pharmacy Technician | 2017-2018
- Dr. Ilia Mazin: PhD, the Ohio State University | 2016-2017
- Dr. Nazar Dubchak: Anesthesiology resident, Walter Reed | 2016-2017
- Jena Jacobs: DO medical student, A.T. Still University | 2015-2017
- Dr. Jessica (Daniel) Watson: MD, University of Florida (Published!) | 2015-2016
- Susan Jett: Biobank Manager, Thermo Fisher (Published!) | 2015-2016
- Aviva Bulow: Software engineer, Sigray | 2014-2016
- Dr. Ryan Warren: Spectrum Health Lakewood, Michigan | 2015-2015
- Dr. Tiffany (Ashbaugh) Tesmer: DO, Midwestern University College of Osteopathic Medicine | 2015-2015
- Michael McCoy | 2015-2015
- Ebony Miller: Certified Nursing Assistant at University of Colorado Health Sciences Center | 2015-2015
- Laura Roon: PRA at Skaggs School of Pharmacy (Published!) | 2014-2015
- Jonathan Richards: Freelance Tutor (Published!) | 2013-2015
- Becky Addison: GIS Software Developer at USGS | 2013-2014
- Jeremy O’Brien: Canning Line Operator | 2013-2014
- Travis Ingraham: Director of Extraction and Quality Assurance for GCH USA | 2013-2014
- Sarai Graves: Provider Relations at United Health Group | 2013-2014
- Kathryn (Norquest) Vang: M.S.; PRA, National Jewish, Colorado (Published!) | 2012-2014
- Dr. Stephen Schaffner: DO, Rocky Vista Medical School (Published!) | 2012-2014
- Kyra Brandt: 1st Lieutenant Signal Officer, US Army | 2012-2014
- Elina (Baravik) Wegner: Oncology R&D at Invitae (Published!) | 2012-2014
- Yerelsy Reyna: Surgeon’s Assistant at Children’s Hospital (Published!) | 2012-2014
- Dr. Josh Sowick: MD, Rosalind Franklin University of Medicine and Science | 2012-2014
- Jody Stephens: Group Lead Software Testing at ArcherDx (Published!) | 2011-2014
- Dr. Ryan Masterson: Resident doctor, Johns Hopkins (Published!) | 2012-2013
- Mason Preusser: Consultant at D-A Lubricant | 2012-2012
- Dr. Tonya Santaus: PhD, University of Maryland Baltimore County | 2012-2012
- Amanda Faux | 2012-2012
- Matthew Stoddard: Scientific Informatics Consultant at Accenture | 2011-2012
- Morgan Miller: Digital Strategy Director at DMA Digital | 2011-2012
Nico Tenorio

Lab Member 2023-2024
Published
Coming soon.
Victoria Colling

Lab Member 2022-2024
Published
Heart disease is one of the leading causes of death in the United States. The lack of effective treatment options and the shortage of available heart donors has led to extensive research investigating new methods to support the structure and function of myocardium tissue. Specific areas of key interest include improvements in transplantation of tissue, human tissue culturing, and the production of extracellular environments in vivo. One of the greatest challenges is improving methods to better mimic the properties of the natural cell environment, such as binding sites, stiffness, reactivity, and hydration. In this research, a conductive polymer matrix will be developed and characterized, which will serve as a scaffold to support the culturation of cardiac spheroids to be used in academics, research, and in medicine. Gold nanoparticles will be synthesized at high purity and with defined surface functionalization, then integrated into gel polymers to provide high conductivity and further support cell retention, spreading, development of cardiomyocytes, and effective gap junctions. Cardiac spheroids will be developed and analyzed using atomic force microscopy (AFM), then introduced into the scaffold. The introduction of these scaffolds with conductive gold nanoparticles into the process of tissue culturing should better enable effective cardiac spheroid maturation. This work will thus advance cardiac tissue engineering and provide methods of determining treatment effectiveness, contributing to improvement in methods for repairing damaged heart muscle and restoring cardiac function.
Keaton Silver

Lab Member 2022-2024
Published
Carrion’s disease is a neglected tropical infection caused by the bacterium Bartonella bacilliformis. Native to South America and transmitted into human vectors via certain phlebotomine species, infection by B. bacilliformis includes both an acute phase of symptoms, such as fever, hemolytic anemia, and myalgia, in addition to a chronic phase, which triggers the proliferation of endothelial cells, often resulting in blindness, and/or skin lesions in the form of Peruvian warts. Indeed, the acute phase of Carrion’s disease can be fatal if undetected and left untreated, making early detection of infection by B. bacilliformis in humans an essential effort. However, most current methods employed in the detection of Carrion’s disease display low sensitivity and require lengthy, expensive workups in a well-outfitted lab setting. Electrochemical DNA-based (E-DNA) biosensors have proven to be rapid, portable, and effective in their ability to detect the presence of small molecule targets through binding to DNA aptamers specific to their target of interest. Therefore, to address the critical challenge of detection, we have selected an aptamer specific to Pap31, a protein found on the extracellular matrix of B. bacilliformis. This aptamer is being adapted to a biosensor format via guided truncation to a minimally active aptamer, then incorporation into a DNA structure that will change conformation upon binding Pap31. This conformation change is turned into an electrochemical voltammetric readout due to the incorporation of methylene blue, a redox-active tag. This will allow the transduction of Pap31 binding into an electrical current signal, and we are verifying the sensitivity and specificity of this biosensor approach. The successful development of an E-DNA biosensor that is fast, effective, and portable for the detection of Carrion’s disease would offer new possibilities for treatment and health outcomes in many South American communities.
Audi Fineran

Lab Member 2022-2023
Published!
Nontuberculous Mycobacteria, referred to as NTM, is an opportunistic pathogen that is becoming a growing concern. NTM causes NTM Pulmonary Disease which often presents similarly to Tuberculosis infections. The current diagnostic “gold standard” for NTM Pulmonary Disease is a microbial culture-based method, which is a weeks-to-months long process. To expedite patient diagnosis and care, we propose a new efficient diagnostic method using a novel electrochemical biosensor that would efficiently detect the presence of NTM. The target of this biosensor is GPL, which is an NTM-specific glycopeptidolipid found within the cell walls of the prominent NTM species. The biosensor will use an electrode-bound DNA aptamer that conformationally changes when GPL binds to it. The conformational change is measured by using voltammetric analysis, which will show a difference when GPL, and therefore NTM, is present. By developing this sensor, it can be utilized medically, environmentally, and ultimately lead to a sensitive and rapid detection of NTM.
Aidan Quinn
Dylan Poch

Lab Member 2019-2022
Current: PhD student, Yale University
Published!
Mycobacterium Tuberculosis (TB) is one of the world’s most prevalent bacterial pathogens. It is estimated that almost 10 million cases of TB emerge every year, and roughly one-fifth of these cases are fatal. The current detection and diagnosis of TB is done primarily via two methods; the TB skin test and the TB blood tests. Neither of these tests can differentiate between latent TB infection and TB disease. In order to differentiate these states, time-consuming sputum tests are required, which rely on culturing the mycobacterium. Designing a sensitive serologic biosensor would dramatically decrease the time line of diagnosis and therefore improve patient outcomes. One possible avenue for improved detection lies in the cell wall of TB, which includes many complex glycolipids—many of which are believed to have immunopathogenic mechanisms in physiologic pathways. Mannose-capped lipoarabinomannan (ManLAM) is one of the most prevalent of these glycolipids, and presents a novel target as a bio-marker for the sensitive detection of TB and related Mycobacterium strains. Here, we have utilized an existing aptamer sequence that binds to ManLAM to generate a sensitive electrochemical, DNA-based biosensor for the detection of TB. This biosensor is able to adopt multiple different folded conformations, only one of which presents the core aptamer sequence in a state capable of binding ManLAM. An appended redox-active tag (methylene blue) generates a measurable difference in electrochemical current upon this conformational change, providing a sensitive and quantitative measurement of ManLAM concentration. Such biosensors may ultimately allow rapid, on site, diagnosis of TB infection within the time constraints of patient-doctor interaction.
[mejsvideo src=”https://www.bonhamlab.com/wp-content/uploads/2021/04/Poch_Biosensors_Spring2021.mp4″ width=”480″ height=”270″]
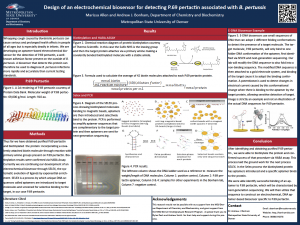
Marissa Allen

Lab Member 2020-2022
Current: PhD student, Marquette University
Whooping cough caused by Bordetella pertussis can cause serious and prolonged health effects, particularly in infants. Quick and accurate diagnosis is essential in treating the infection. Early treatment can significantly reduce the duration of illness in the patient and lead to a milder case of illness overall. The current standard for testing set forth by the CDC is invasive, requiring nasopharyngeal swabs. Most medical facilities are not equipped to then process the sample collected, and it is instead sent out for analysis adding to the length of time required to get a diagnosis for the patient. Common laboratory tests for the detection of B. pertussis are PCR and culture, but each present unique challenges. The purpose of our research is to design and develop a rapid electrochemical biosensor that can detect P.69 pertactin. P.69 pertactin is a surface protein used by B. pertussis to bind to mammalian cells. A biosensor that can detect P.69 pertactin would improve several major issues associated with the current testing standards including size of the patient sample needed, time to get results, cost of testing and shipping, and accuracy of tests. The B. pertussis biosensor will be developed through the SELEX process and will create a specific oligonucleotide probe for P.69 pertactin . This probe will then be adapted into an electrochemical DNA-based biosensor. This biosensor would require a much smaller sample from the patient and give almost immediate results to the physician overseeing the case.
Mary Quansah

Lab Member 2021-2022
Published!
Human Ecto-NOX Disulfide-Thiol Exchanger 2 (ENOX2) is a cell-surface metalocatalyst which is expressed solely on malignancies of all major human cancer types. ENOX2, along with its ubiquitous parent protein ENOX1, serve as terminal oxidases in plasma membrane electron transport chains, suggesting that such proteins are essential for cell growth. ENOX2 is frequently shed from malignancies into the sera and demonstrates site-specific transcript variants, and thus presents an attractive target for both diagnostic blood testing and anti-cancer therapeutic treatments. To better facilitate detection and targeting of ENOX2 in patient sera, we utilized SELEX to generate aptamers against recombinantly expressed ENOX2. These aptamers were characterized via gel mobility shift assays to determine the candidate aptamer with highest affinity for human ENOX2. The candidate with highest affinity was incorporated into an electrochemical DNA-based biosensor, where changes in DNA confirmation upon target binding result in changed dynamics of an appended redox reporter molecule, ultimately generating a rapid and quantitative change in the current of the electrochemical response. This novel ENOX2-targeting electrochemical biosensor allows rapid, dose-responsive readout for the presence of ENOX2 in both buffer and blood serum.
Chris Chamblee

Lab Member 2020-2022
Current: PhD student, Texas A&M
The goal of this project is to develop an electrochemical biosensor capable of rapid detection of pathogenic Mycoplasma bacteria in academic, research, and clinic settings. This research is highly significant because this biosensor will be capable of detecting two of the three pathogenic Mycoplasma strains that cause human disease, namely Mycoplasma pneumoniae and Mycoplasma genitalium. Mycoplasma pneumoniae, the more prevalent of the two, infects almost two million people every year in the U.S. It is responsible for contagious upper respiratory infections and is also known as atypical or “walking pneumonia” that spreads rapidly in crowded areas2. The second most prevalent, Mycoplasma genitalium, is responsible for approximately 15%–20% of nongonococcal urethritis (NGU) cases, 20%–25% of nonchlamydial NGU, and approximately 30% of persistent or recurrent urethritis3. In addition to causing these human diseases, Mycoplasma also continues to be a major contaminant of laboratory cell cultures4. Current methods of detections for Mycoplasma bacteria, such as molecular-based assays, PCR and serological analysis are time consuming, expensive, and less suitable for working under stringent conditions such as extreme temperature or pH1. Moreover, current serological methods create possibilities of false negative results for most infected individuals because they do not measure the presence of the microorganism. They instead focus on measuring the host immune response. Therefore, our research will aim at detecting the presence of these Mycoplasma strains with a diagnostic electrochemical biosensor that is designed for rapid, reliable, and reagentless serological detection of several common Mycoplasma strains. Using this electrochemical biosensor we will be able to detect each listed pathogenic Mycoplasma, by identifying a commonly secreted protein, P48. Dr. Bonham and his past student researchers designed a successful aptamer (a DNA oligonucleotide designed to selectively bind to and detect the P48 protein) with which we will work to create a final product that is sensitive enough to detect P48 protein in a minimal amount of human serum. This will allow us to potentially improve both prevention and diagnosis of Mycoplasma in patients who present a proposed infection.
[mejsvideo src=”https://www.bonhamlab.com/wp-content/uploads/2021/04/Chamblee_Biosensors_Spring2021.mp4″ width=”480″ height=”270″]
Andrew Smith

Lab Member 2019-2021
Current: MD Student, Kansas University
Published!
Carrion’s disease is a neglected tropical disease (NTD) caused by infection by the bacteria B. bacilliformis, endemic to northern Peru, and affects primarily rural, impoverished populations. In rural areas, diagnosis is currently made using Giemsa-stained blood smears, but this technique has a very low sensitivity for the disease (24-36%) and requires trained individuals, which are in short supply. The disease can be diagnosed via qPCR, IFA, and ELISA tests, and show high sensitivity, but are not feasible for diagnosis in rural areas, due to their impoverished nature. Here, we propose start-up activities that should allow the creation of a biosensor-based strategy for field detection of the bacteria responsible for CD. Our CD biosensor will be developing through a process known as SELEX: systematic evolution of ligands by exponential enrichment using the commercial x-aptamer selection system. This process will generate a specific probe against the Pap31 protein shed by the pathogenic bacteri responsible for CD which will be adapted into an electrochemical DNA-based biosensor. E-DNA sensors should provide a rapid means of testing for CD. These sensors have a long shelf-life and are reusable, as well as requiring only small blood samples. Electrochemical sensors provide easily read and interpreted output suitable for point-of-care conditions and can be analyzed using portable equipment on site. Thus, only a finger lancet-derived blood sample obtained in the field would be required. Ultimately, these characteristics offer the broader impact of better, timelier and less invasive diagnostics for those affected by this disease.
[mejsvideo src=”https://www.bonhamlab.com/wp-content/uploads/2021/04/Smith_Biosensors_Spring2021.mp4″ width=”480″ height=”270″]
Dr. Tyler Sodia

Lab Member 2018-2020
Current: Particle Scientist, VitriVax; PhD, Colorado School of Mines
Published!
Mycobacterium Tuberculosis (TB) is one of the world’s most prevalent bacterial pathogens. It is estimated that almost 10 million cases of TB emerge every year, and roughly one-fifth of these cases are fatal. The current detection and diagnosis of TB is done primarily via two methods; the TB skin test and the TB blood tests. Neither of these tests can differentiate between latent TB infection and TB disease. In order to differentiate these states, time-consuming sputum tests are required, which rely on culturing the mycobacterium. Designing a sensitive serologic biosensor would dramatically decrease the time line of diagnosis and therefore improve patient outcomes. One possible avenue for improved detection lies in the cell wall of TB, which includes many complex glycolipids—many of which are believed to have immunopathogenic mechanisms in physiologic pathways. Mannose-capped lipoarabinomannan (ManLAM) is one of the most prevalent of these glycolipids, and presents a novel target as a bio-marker for the sensitive detection of TB and related Mycobacterium strains. Here, we have utilized an existing aptamer sequence that binds to ManLAM to generate a sensitive electrochemical, DNA-based biosensor for the detection of TB. This biosensor is able to adopt multiple different folded conformations, only one of which presents the core aptamer sequence in a state capable of binding ManLAM. An appended redox-active tag (methylene blue) generates a measurable difference in electrochemical current upon this conformational change, providing a sensitive and quantitative measurement of ManLAM concentration. Such biosensors may ultimately allow rapid, on site, diagnosis of TB infection within the time constraints of patient-doctor interaction.
Lindsay Armstrong
Electrochemical biosensors based on the conformational dynamics of DNA aptamers have found success against a wide variety of proteins, toxins, antibodies, and heavy metals. However, the mechanistic underpinnings of the mechanism by which these surface-bound DNA molecules change conformation upon target binding, thus changing the dynamics of an appended redox-active tag and generating a measurable signal, is poorly understood. Our first target for investigation was a previously reported Ricin Chain A binding atamer biosensor. Here, we have investigated this biosensor using a variety of nucleic acid assays, including PCR-termination via basepair modification, fluorescence anisotropy, gel mobility shift assays, and FRET tagging to determine 3D orientation. These have allowed us to better characterize the basepair interactions involved in binding targets, as well as offer clues to the changing three dimensional folded structures of these biosensors. These results will help inform the field of biosensors and aptamers in general on strategies for future optimization.
Austin Haider
Current: PhD student, Denver University
Lab Member 2018-2019
Project: Algorithmic Prediction of DNA Biosensor Structures
Oligonucleotide-based biosensors have been demonstrated as effective tools for detecting heavy metals, small molecule drugs, and protein targets. Recent efforts to move these biosensors from the lab bench to practical use in industry and medicine rely on the ability to rapidly and effectively adapt them to detect new targets of interest. One compelling aspect of DNA and RNA-based biosensors is the great strides that have been made in computational prediction of the three-dimensional structures that they can assume. However, interpreting predicted structures and using that knowledge to design functional biosensors ab initio remains a challenging problem. We hypothesize that by representing a potential DNA biosensor as a node-weighted graph, we can simplify the challenge of automated structure interpretation and sorting. If correct, this would allow a researcher to provide a core sequence of interest (such as a DNA recognition element or artificially-selected aptamer) and have a software tool design the ideal DNA biosensor for the sensitive detection of that target. This process builds on ideas from our oligionucleotide fold scoring algorithm, Fealden. With this new implementation of Fealden, we are achieving significantly more flexibility, allowing us to analyze any structure type. Because of this we are able to traverse a search space of tens of thousands of possible sequences to determine a “best candidate” biosensor. Additionally, with this increased flexibility we hope to implement some very desirable features in our software, generally, different types of design constraints that a researcher can specify for their biosensor.
Marcos Maldonado

Lab Member 2016-2019
Published!
Cardiomyopathies, diseases of the heart, are one of the major causes of death in the United States, and thus there is great interest in preventing and treating these complications. However, due to the nature of limited availability of donors, many techniques and solutions are inadequate to meet the needs of the field. In particular, heart tissue transplantation, culturing human tissues/tissue-derived cells and tissue engineering, and finding a suitable extracellular environment that closely resembles the natural environment of cardiomyocytes in vivo are difficult to attain. As such, a great deal of work has gone into efforts to produce polymers which mimic the natural cell environment in properties such as binding sites, stiffness, reactivity, and hydration. In our research, we are investigating the development and characterization of conductive polymer scaffolds providing cardiac tissue support, which will ideally aid in culturing cardiomyocytes for academic, research, and medical use. The core of our model is the incorporation of conductive gold nanorods into reverse thermal gel polymers. Synthesized gold nanorods utilized in this procedure are made with a high aspect ratio, at high purity and with defined surface functionalization. These conducting scaffolds should properly accommodate cardiac cells in building functional cardiac tissue constructs and further improving cell retention, spreading, homogenous distribution of cardiac specific markers, cell-cell coupling and synchronized beating behavior at the tissue level. The nanoparticles have been synthesized, then covalently coupled into these scaffolds, resulting in order-of-magnitude increases in conductivity. The goal of this work aims to improve cardiac tissue engineering, so that it can be directed to ultimately repairing damaged heart muscle and improve overall cardiac function in cardiovascular diseases whether they have been acquired from past medical complications or developed through hereditary traits.
Anika James

Lab Member 2018-2019
Current: PhD student, Kansas University
Mycoplasma bacteria are highly infectious agents of human disease and laboratory contamination. For example, Mycoplasma pneumonia infects almost two million people every year with contagious upper respiratory infections also known as atypical or ‘walking pneumonia’. In addition to human disease, Mycoplasma is a major source of contamination of laboratory human cell cultures. However, current methods of detection for Mycoplasma strains, such as molecular-based assays, PCR and serological analysis, are time consuming, expensive, and less suitable for working under stringent conditions such as extreme temperature or pH. To provide a rapid and accurate alternative, we have focused on detecting the presence of Mycoplasma with a diagnostic electrochemical biosensor that detects lipoprotein P48, which is shed from the surface of several strains into the surrounding blood serum or growth media. This biosensor is based on a previously identified DNA aptamer that binds to the secreted P48 protein with high affinity. We have expressed P48 recombinantly in E. coli to serve as a positive control, and our results show rapid and sensitive binding of the target, as well as convenient electrochemical signaling from these binding events. This approach will allow us to potentially improve both prevention and diagnosis of Mycoplasma in cell culture platforms as well as patients who present a proposed infection.
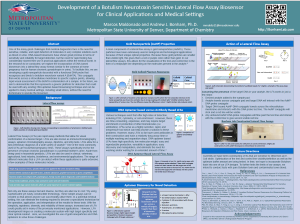
Marlea Kudlauskas

Lab Member 2018-2018
Current: Cardiac ICU Nurse
One of the many great challenges that medical diagnostics face is the need for sensitive, reliable, and rapid detection of molecules in very complex solutions such as blood or urine. DNA-based biosensors have shown great promise in terms of sensitivity and reliability for target detection, but the need for rapid testing has considerably slowed their use in practical applications within the medical world. In the research to be conducted, we explore the incorporation of DNA-based biosensors into a lateral flow assay format (similar to the common at-home pregnancy test for human chorionic gonadotropin in urine). To facilitate this, we are developing a gold nanoparticle decorated with a functional DNA probe that recognizes and binds to botulism neurotoxin variant A (BoNTA). This conjugate then wicks across a nitrocellulose membrane to specific capture points, allowing rapid visual assessment of the BoNTA contamination of a sample. In the future, we aim to demonstrate that this represents a generic platform for detection that could be used with any existing DNA aptamer-based biosensing technique and can be applied to many medical settings, including small clinics, without the need for technicians to operate the biosensor.
Anna Nguyen

Lab Member 2017-2018
Current: PhD student, University of California Santa Barbara
Published!
Celiac disease is an autoimmune disorder where ingesting dietary gluten causes an autoimmune response that leads to villous atrophy and other complications. Symptoms can have gastrointestinal or extraintestinal manifestations. The symptoms can include: diarrhea, abdominal pain, dental enamel defects, neurological issues, Dermatitis Herpetiformis and more. This disease affects an estimated 2 million Americans, 1.4 million of which are undiagnosed1. Rate of diagnosis is low because many affected individuals have “silent celiac disease” where the symptoms are not detectable, but villous atrophy is still occurring. Also, there is a broad spectrum of symptoms for the condition and is often misdiagnosed. The average time it takes to diagnosis individuals who are symptomatic in the US is 11 years2. In that time, their risk of developing other autoimmune disorders, neurological problems, fertility problems and other conditions increases. One key challenge of this condition is that the diagnosis has historically suffered from difficulties in performing non-invasive positive identification of affected patients. Recently, interacting amino acid sequences on the surface of human tissue transglutaminase and gliadin proteins from foodstuffs have been identified as the potential recognition sites of commonly developed auto-antibodies in Celiac disease. In this project, we are exploring the use of short synthetic peptides that mimic these key amino acid recognition sites in order to create rapid, non-invasive, electrochemical biosensors to aid in the early diagnosis of Celiac disease. These biosensors build on a literature of electrochemical biosensors that offer reagentless, reusable, and rapid testing directly in small quantities of blood (such as a finger lancet draw). Our goal is to design, synthesize, and validate such diagnostic biosensors.
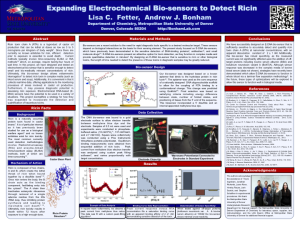
Lisa Fetter

Lab Member 2014-2018
Current: PhD student, University of California Santa Barbara
Published!
Ricin toxin chain A (RTA), a byproduct of the production of castor oil from castor bean plants, is a hazardous toxin that inhibits the cellular production of proteins once it enters the body. This toxin, whether ingested, inhaled, or injected, can be lethal, and treatment is difficult as there are currently no known antidotes for ricin. Detection of RTA prior to exposure is thus important, and it is necessary to expand the methods that can be used for this detection, as current methods are not time-sensitive. In this project, we have designed and tested an electrochemical biosensor that is capable of– and sensitive enough– to detect small, bio-medically relevant concentrations of RTA. This biosensor was designed based on an existing oligonucleotide aptamers that have been previously shown to bind to the hydrolase protein in ricin toxin. One of these aptamers was then used as the basis of a rationally designed DNA oligonucleotide biosensor scaffold that allows the coupling of RTA binding to a conformational change in the oligonucleotide. Ultimately, this biosensor design allows voltammetric interrogation to detect RTA concentration in complex media (such as coffee, blood, and river water). Additionally, this biosensor is convenient and collects real-time data, offering beneficial applications to the monitoring processes of areas involved in castor oil production. Furthermore, it may possess diagnostic potential in assessing ricin exposure. Electrochemical DNA biosensors have the potential to be used in numerous different situations, and this project shows a strategy for how they may be expanded to the detection and quantification of hazardous toxins.
Derek Clark

Lab Member 2017-2018
Current: Pharmacy Technician
Gold nanoparticles, typically particles with a diameter of < 100 nanometers, present unique electrical and optical properties from the bulk material. These properties have been used to enable a variety of sophisticated molecular and chemical techniques. One of the most intriguing of these is the phenomenon of surface-enhanced Raman scattering, wherein closely spaced nanoparticles dramatically increase the sensitivity of Raman, a technique that allows “fingerprinting” of molecules. In this project, we synthesized gold nanoparticles, coated them with single stranded thiolated DNA. In the near future, we will anneal two variations of functionalized gold nanoparticles together. A functionalized Gold nanoparticle scaffold will form and promote signal Raman amplification due to calculated spacing. Designated proteins will bind to specific regions of DNA in the scaffold. binding should alter the molecular fingerprint of an organic dye, methylene blue, that is attached to the DNA. Ultimately, such assemblies should offer a stable optical platform for the creation of incredibly sensitive biosensors.
Dr. Ilia Mazin

Lab Member 2016-2017
Current: PhD, the Ohio State University
Building off of existing electrochemical biosensors that have been developed by the Bonham lab, this research aims to adapt these sensors’ responses when recognizing their target from electrochemical to be visual in nature. Namely, this biosensor consists of a 3D-printed brace, an acrylic slide on which DNA is immobilized, and readily-available electronics: an inexpensive LED light source and a cylindrical lens to direct the light in an elliptical pattern. The scaffold allows light to enter the slide at above the critical angle, for total internal reflectance, ensuring that the immobilized DNA experiences excitation by evanescent wave. If the immobilized DNA is an aptamer-based biosensor with a fluorescent reporter tag, binding of target that is washed across the slide surface will result in characteristic and measurable fluorescence changes. The utility of this inexpensive DIY setup was evaluated with image-analysis software to create dose-response curves for the response of a protein-binding aptamer for its target and demonstrates the potential utility of these evanescent-wave biosensors.
Dr. Nazar Dubchak

Lab Member 2016-2017
Current: Anesthesiology resident, Walter Reed
B-type natriuretic peptide (BNP) is a commonly used biomarker for the onset of heart failure conditions, as its secreted levels in the blood correlate with internal pressure of cardiac tissue, and high levels of BNP are typically found in patients with congestive heart failure. Unfortunately, current tests for BNP are largely based on phlebotomic blood draw followed by ELISA, which requires external testing and eliminates the possibility of point-of-care diagnosis. To address this unmet need, we have developed a rapid, convenient biosensor for BNP based on a DNA aptamer specific to BNP that has been developed into an electrochemical readout format. Such electrochemical DNA (E-DNA) biosensors have been shown to function in unprocessed whole blood and yield quantitative results in less than one drop of blood, making a finger lancet-based, point-of-care assay possible.
Jena Jacobs

Lab Member 2015-2017
Current: DO medical student, A.T. Still University
Mycoplasma pneumonia infects 2 million people every year and is responsible for upper respiratory infections and “walking pneumonia.” Here, we describe the creation of a novel electrochemical biosensor capable of detecting pathogenic Mycoplasma for use in academic, research, and clinical applications. Current diagnostics of Mycoplasma, such as molecular-based assays, PCR and serological analysis, are time consuming, expensive, and not particularly accurate. In response, our biosensor is designed for rapid, reliable, and reagentless detection of several common Mycoplasma strains. To do so, we rely on the fact that many pathogenic Mycoplasma share a common secreted protein, P48. A modified aptamer against P48 was incorporated into a custom oligonucleotide scaffold and is used in a gold-electrode-bound fashion to give electrochemical signal change upon binding the secreted P48 target. Ultimately, this biosensor should bring improvements to diagnosis and thus treatment of Mycoplasma in patients who present a proposed infection.
Dr. Jessica (Daniel) Watson

Lab Member 2015-2016
Current: MD, University of Florida
Published!
Although rationally designed biosensors often perform well in buffered media under lab bench conditions, that is insufficient for real-world performance. Thus, I focus on using small sample volumes, complex media, and rapid (<15 minutes) analysis, to make biosensors suitable for point-of-care and inspection use. This work may include changing surface passivation, pretreatment, surface protection and filtering, and creating software front-ends that rapidly evaluate results and interpret the data into actionable metrics.
Susan Jett

Lab Member 2015-2016
Current: Biobank Manager, Thermo Fisher
We have developed a novel electrochemical biosensor for the purpose of detecting uranium, in the uranyl ion form, in untreated water samples at low levels for use in academic, research, and professional settings. This could lead to applications in ongoing water quality monitoring, as well as improve targeting and efficiency of remediation of contaminated water sources. Uranium contamination of a water source has far reaching implications as a connected water system allows for contaminates to pollute all connected water systems over many miles, in turn effecting the organisms that rely on that water supply. This system uses a modified uranyl-binding aptamer that has been inserted into a oligonucleotide scaffold to create a conformation-switching element. This oligonucleotide is then integrated into an electrochemical biosensor platform. Ongoing work is determining the detection limits and sensitivity to the target ion of our scaffold biosensor, with the goal of parts per billion detection of uranyl, allowing compliance measurement against EPA standards of uranium limits.
Aviva Bulow

Lab Member 2014-2016
Current: Software engineer, Sigray
Oligonucleotide-based biosensors have been demonstrated as effective tools for detecting heavy metals, small molecule drugs, and protein targets. Recent efforts to move these biosensors from the lab bench to practical use in industry and medicine rely on the ability to rapidly and effectively adapt them to detect new targets of interest. One compelling aspect of DNA and RNA-based biosensors is the great strides that have been made in computational prediction of the three-dimensional structures that they can assume. However, interpreting predicted structures and using that knowledge to design functional biosensors ab initio remains a challenging problem. We hypothesize that by representing a potential DNA biosensor as a node-weighted graph, we can simplify the challenge of automated structure interpretation and sorting. If correct, this would allow a researcher to provide a core sequence of interest (such as a DNA recognition element or artificially-selected aptamer) and have a software tool design the ideal DNA biosensor for the sensitive detection of that target. This process builds on ideas from our oligionucleotide fold scoring algorithm, Fealden. With this new implementation of Fealden, we are achieving significantly more flexibility, allowing us to analyze any structure type. Because of this we are able to traverse a search space of tens of thousands of possible sequences to determine a “best candidate” biosensor. Additionally, with this increased flexibility we hope to implement some very desirable features in our software, generally, different types of design constraints that a researcher can specify for their biosensor.
Dr. Ryan Warren

Current: Spectrum Health Lakewood, Michigan
Lab Member 2015
Metallic nanoparticles (NP) have been used as the basis of versatile new classes of bio-medical tools. NP-biopolymer conjugates have been used as sensitive tumor markers, MRI imaging contrast enhancement tools, novel chemotherapeutics, and as biosensors. Particularly, NP biosensors can be built that use the unique surface plasmon features of metallic NP to enhance the sensitive molecular fingerprinting technique Raman spectroscopy. Here, we aim to expand on the utility of rationally designed DNA oligonucleotide-based biosensors by expanding them from electrode-bound platforms to NP-conjugated, solution-phase systems. Gold nanoparticles (10-50 nm diameter) were synthesized via citrate reduction of gold aurate, followed by conjugation to thiol-modified DNA probes and spectroscopic characterization. The NP-DNA conjugates display unique molecular fingerprints, and we hypothesize that these unique spectroscopic features can be used to track the association of small molecule and protein targets of interest with the NP-DNA assemblies. This would provide a rapid, responsive measure of target concentration using Raman spectroscopic investigation. Field portable Raman tools are already common in the manufacturing and food safety industries, and those tolls, with our NP-DNA assemblies, could be adopted rapidly into a useful new class of medical point-of-care diagnostic. This work describes our fabrication and characterization process, as well as our results using the NP-DNA platform to detect transcription factor proteins.
Dr. Tiffany (Ashbaugh) Tesmer
 Lab Member 2015-2015
Lab Member 2015-2015
Current: DO, Midwestern University College of Osteopathic Medicine
Many modern protein analytical characterization techniques, such as X-ray crystallography, nuclear magnetic resonance, or activity gel shift assays rely on access to large quantities of purified human proteins that have been recombinantly expressed in bacteria. In particular, our lab builds electrochemical and optical biosensors to detect proteins involved in cancer that require pure recombinant proteins for validation purposes. However, the process of expressing and purifying functional human proteins in bacteria requires optimization on a protein-by-protein basis. Here, we investigate varied buffer parameters in the process of affinity column chromatographic separation of recombinant Max (myc-associated factor X) transcription factor protein. The gene encoding a version of Max that is fused to a his-tag purification motif was previously introduced into BL21 E. coli bacteria, and this bacteria were used to produce large quantities of cell lysate. However, the process of isolating Max from the thousands of native bacterial proteins requires careful selection of purification procedures. In particular, the concentration of imidazole, an amino acid mimic used to out-compete Max binding to the affinity column, has dramatic effect on the final purity of the protein obtained. By varying the concentration of imidazole, we aim to allow efficient and optimized purity of Max for testing of our lab’s biosensors.
Michael McCoy
Ebony Miller

Lab Member 2015-2015
Current: Certified Nursing Assistant at University of Colorado Health Sciences Center
Electrochemistry is an industrially, analytically, and medically important field of chemistry; however, electrochemistry concepts and techniques are rarely introduced in the general chemistry curriculum. Two factors that have contributed to this lack of incorporation are one, that electrochemistry is widely viewed as non-intuitive, requiring additional physics course preparation, and two, because reliable electrochemistry instrumentation has remained outside the budget of student laboratories. In this work, introductory electrochemistry experiments are being investigated for incorporation into the general chemistry curriculum. The guided-inquiry lab experiences will utilize a low-cost electrochemistry device—the CheapStat—that Dr. Bonham et al. have previously developed, allowing us to develop self-guided labs that use instrumentation that can be provided to individual students instead of using limited time on a single classroom machine. These experiments will use electrochemistry to investigate relatable concepts such as verifying the concentration of pain relievers in common cold medicine. It is hypothesized that students who complete these self-guided labs will gain a deeper insight into electrochemical theory and techniques. Currently, labs are being developed and verified to ensure they are reliably reproducible, engaging, and incorporate best practices for self-guided lab experiences. In the future, we plan to assess general chemistry laboratories that complete or do not complete these modules and assess the impact that they make on the educational experience and knowledge of electrochemistry.
Laura Roon

Lab Member 2014-2015
Published!
DNA-based electrochemical biosensors offer a unique method for the selective detection of protein. We have developed and optimized a sensitive, electrochemical biosensor to detect the presence and concentration of the transcription factor c-Myc. c-Myc plays key roles in regulating many aspects of cellular replication and growth; as a consequence this transcription factor is crucially misregulated in many breast and kidney cancers. Several studies have identified a correlation between elevated intracellular levels of c-Myc and the presence of cancerous tissue. Although prior oligonucleotide-based biosensors against c-Myc have been developed, they rely on fluorescence spectrophotometers– equipment generally unavailable in point-of-care applications. In response, we have expanded this detection into an electrochemical format suitable for use with portable instrumentation, small volumes, and pre-fabricated patterned microelectrodes. This biosensor has the potential to serve as an affordable, quantitative, and point-of-care tool for clinical diagnosis and academic research. Here, we demonstrate their function on conventional and micro-electrodes in buffer as well as their function in crude samples (e.g., blood serum), achieving sensitive and quantitative detection of c-Myc. These designs can be rationally extended to detect other transcription factors of interest. Thus, this class of electrochemical biosensors is poised to serve as potent point-of-care diagnostics in biomedical applications.
Jonathan Richards

Lab Member 2013-2015
Published!
[mejsvideo src=”https://www.bonhamlab.com/wp-content/uploads/2013/12/Richards_URC2014_web.mp4″ width=”480″ height=”270″]Botulinum neurotoxin serotype A (BoNT/A) is a dangerous substance produced by the anaerobic bacteria Clostridium botulinum, which causes human botulism. Common infection sources include IV contamination, food sources and soil. The complexity of food and blood samples where BoNT/A contamination is suspected makes detection difficult compared to purified laboratory samples. Standard detection methods for botulinum neurotoxins require at least a day for results and have up to a ~30% failure rate.3 These lengthy diagnostics mean that doctor examination if often the primary means of diagnosis. The creation of sensitive, real-time DNA aptamer-based electrochemical biosensors to detect BoNT/A could significantly help alleviate these challenges in diagnosis and saftey. Such BoNT/A biosensors could be used for detection of botulinum toxin in presumptively infected individuals and in food production facilities, leading to rapid diagnosis of botulism and better detection of BoNT/A contaminated products. In order to facilitate the creation of these biosensors, we have used a selection process called SELEX (Systematic Evolution of Ligands by Exponential Enrichment) to discover a novel DNA-based aptamer that will bind to an atoxic derivative of BoNT/A. Aptamers are synthetic oligonucleic acids that bind to a specific protein target, and current efforts have yielded a pool of potentially sensitive aptamers. Next generation DNA sequencing, sequence alignment studies, and magnetic bead pulldown assays will be used to identify the best biosensor candidate. Future efforts will incorporate these BoNT/A aptamers into the existing E-DNA electrochemical biosensor platform.
Becky Addison

Lab Member 2013-2014
Current: GIS Software Developer at USGS
Human transcription factors are proteins that play an important role in both normal development and in many disease responses. Recent bio-engineering advances have led to a specific design of a structure switching fluorescent bio-sensor which can detect a specific transcription factor. This bio-sensor utilizes a sequence of DNA that can fold into two different structures in near equilibrium. However, there are thousands of human transcription factors, and we have thus generalized this design and implemented a software platform, Fealden, which generates sensors for any transcription factor. In this platform, we use a depth first search to find a possible sequence. We determine the validity of this sequence by leveraging existing energy minimization software tools to predict all possible conformations of the sequence, then analyzing that output for metrics including point to point distances and stem presentation. Upon completion of this process, a successful bio-sensor design is returned with calculated equilibrium constants and free energy values. Current work is focused on generalizing this platform to work with more complex bio-sensor designs, such as enabling expansion into electrochemical sensors. This involves detailed calculation of distance between arbitrary points in a 3D DNA folding, as well as generalizing how we prune our search tree. Ultimately, this algorithm design will provide a general, expandable representation of these transcription factor sensor designs, providing the ability for academics and medical technicians to more accurately and readily measure transcription factor levels to diagnose and understand cellular processes.
Jeremy O’Brien

Lab Member 2013-2014
Current: Canning Line Operator at Los Bucaneros
The overall purpose of this research project is to design and synthesize a library of organic compounds with the goal of creating potent inhibitors of the Myc-Max transcription factor complex binding to DNA. There will be three separate phases of this project. The first is computational chemistry, using chemical structure design software coupled with molecular dynamics-based ligand binding software (Autodock) to design inhibitory compounds and predict their affinity for Myc-Max in silico. Once likely candidate molecules have been identified, the second focus, synthesis of the likely inhibitors, will commence. Along with applying modern synthetic techniques, success of the newly created compounds will be verified with nuclear magnetic resonance (NMR) spectroscopy and gas-chromatography-mass-spectrometry (GC-MS), both available within the department. Finally, the efficacy of the synthesized inhibitors will be determined using a novel fluorimetric assay based on prior work with ‘TF Beacon’ bio-sensors, which report on protein:DNA interactions in real-time using existing lab expertise and protein reagents.
Travis Ingraham

Lab Member 2013-2014
Current: Director of Extraction and Quality Assurance for GCH USA
Ricin is toxic protein produced in high concentrations in the seed of the castor oil plant, and can be easily extracted by rudimentary means in large quantities. As it has the potential to be used as a bio-terrorism agent, there is a pressing need for methods to quickly detect Ricin in water supplies, foodstuffs, and blood samples. We have developed a sensitive, electrochemical DNA bio-sensor (E-DNA sensor) directed against this target, based on synthetic Ricin-binding aptamer structures. These sensors give an easily measured change in electrical current in response to the concentration of Ricin present in a sample. Ongoing studies are evaluating the efficacy of this detection and the capability for real-world detection in complex media with low concentrations.
Sarai Graves

Lab Member 2013-2014
Current: Provider Relations at United Health Group
Kathryn (Norquest) Vang

Lab Member 2012-2014
Current: Masters student, Notre Dame University
Published!
Retinoblastoma is a destructive childhood cancer that develops in the retina of the eye and often results in blindness. Interestingly, a large portion of children with the disease have a mutation in the gene RB1, which codes for a tumor suppressor protein. The retinoblastoma protein plays a critical role in regulating cell cycle progression by binding members of the E2F family of transcription factors. These are proteins that regulate gene expression. To gain insight into the means by which Rb dysfunction affects the cell, we are investigating recombinantly expressed and purified constructs of Rb and E2F1 to determine the impact of Rb on E2F1 DNA-binding activity. In order to observe this process, we are simultaneously developing sensitive, DNA-based bio-sensors that allow rapid detection of these interactions with a simple fluorescent readout.
Dr. Stephen Schaffner

Lab Member 2012-2014
Current: DO, Rocky Vista Medical School
Published!
A patient’s chances of surviving cancer increase significantly with early detection, and aggressive treatment in the early stages of disease. The long term goal of this project is the creation of a rapid blood test to help improve the odds of early detection. To that end, a clear understanding of the complex dynamics of the myriad molecular factors involved in both normal and cancerous cell growth is necessary. The proper regulation of cellular growth cycles is critical in maintaining the biochemical steady state present in every human cell. The deregulation of transcription factors controlling cell growth and development is a hallmark of malignant tumorigenesis, and this necessity opens the door to the possibility of rapid and early detection of cancers through analysis of transcription factor concentration. Upregulated, mutated, or highly stabilized transcription factor c-Myc is associated with numerous cancers including approximately 25% of breast cancer incidences. This transcription factor preferentially binds with the straightforward DNA sequence “CACGTG”, making it a good candidate for initial research into methods for in vitro concentration analysis. The c-Myc protein also interacts with a number of other associated transcription factors including Max, Miz1 and Sp1, forming heterodimers which have a known impact on the binding affinity and specificity of c-Myc. A better understanding of c-Myc’s binding activity and concentration in different pre- and post-tumorigenic conditions would enable a straightforward screen for early onset of certain cancers.
Kyra Brandt

Lab Member 2012-2014
Current: 1st Lieutenant Signal Officer, US Army
The development and growth of neurons in the human brain is highly controlled to ensure proper maturation. These processes are largely regulated by proteins known as transcription factors that turn on and off specific genes. Interactions between these proteins are known to have large functional consequences, but the detailed kinetic and thermodynamic parameters that ultimately decide cell state remain unclear for many protein systems. This is of particular interest to understand differentiation and genetic disease, such as the mechanisms by which the proteins Ascl1 and Lmo4 direct neuronal commitment. Ascl1 is expressed in high concentrations in cancers such as medullary thyroid and small cell lung cancer and is potentially useful in detecting early stages of these diseases. To explore the common mechanisms by which these systems control gene expression, we have generated sensitive, DNA-based biosensors for the assessment of the DNAbinding activity of these significant proteins. The proteins were cloned in a plasmid, expressed in bacteria, purified, and confirmed on a western blot. Ascl1 binding activity was probed using novel DNA biosensors and found to have an affinity of 108 nM. Investigations have begun on the effects of Ascl1’s binding partner, Lmo4, to understand how these interactions control DNA binding specificities. Using these sensors to investigate recombinant protein systems will provide a greater mechanistic understanding of the effect of these interactions on their regulatory activity and functional consequences in the cell.
Elina (Baravik) Wegner

Lab Member 2012-2014
Current: Oncology R&D at Invitae
Published!
The Encyclopedia of DNA Elements (ENCODE), a project sponsored by the National Human Genome Institute, is a collaborative effort to identify all areas of transcription, transcription factor binding, chromatin structure and histone modification in the human genome. One recent ENCODE study utilized high- throughput CHIP-sequencing and DNase I footprinting to investigate protein- protein interactions across the genome. Of note, this study discovered a large number of novel protein-protein interactions that have never been observed in prior work. As a continuation and validation of this effort, we are characterizing the novel binding interaction identified between ATF3 and MAX, two transcription factor proteins deeply involved in cell regulation and function. These proteins natively bind to DNA, MAX directly and ATF3 indirectly, and we are probing the impact of their interactions with each other on their DNA binding affinity and specificity. To do so, we are using DNA-based bio-sensors that give off fluorescent signal proportionate to the extent of DNA-binding activity. We designed these bio-sensors from first principles, and are planning to express, purify the protein targets, and have begin to investigate their binding interactions. This work will confirm if the proposed physical interaction is indeed valid, and will shed light on the mechanism by which they bind to the DNA and each other. Ultimately, this data will enrich the depth of the ENCODE project and provide a greater understanding of novel cellular pathways involved in cell growth and development.
Yerelsy Reyna

Lab Member 2012-2014
Published!
Transcription factors bind to specific genomic sequences to control the flow of genetic information from DNA to mRNA. The preinitiation complex of these proteins includes TATA binding protein (TBP), which specifically recognizes the TATA box, a highly conserved region at the beginning of many genes. As many cancers result from the aberrant regulation of transcription factors, there is great interest, both academically and diagnostically, in a better understanding these binding events, and TBP is a core factor that can help guide the design of novel probes of transcription. Here, we report on the design and use of DNA-based folding biosensors that are capable of quantitatively and sensitively detecting TBP using custom optical and electrochemical probes that are activated by TBP’s intrinsic DNA-binding activity. These sensors will ultimately allow us to investigate the role that protein-protein interactions and inhibitory drugs have on gene expression.
Dr. Josh Sowick

Lab Member 2012-2014
Current: MD, Rosalind Franklin University of Medicine and Science
This project is concerned with the development, optimization, and evaluation of sensitive, electrochemical biosensors to detect the presence and concentration of the transcription factor c-Myc. c-Myc plays a key role in regulating most aspects of cellular function, including replication, growth, metabolism, differentiation and apoptosis. This sensor will extend the E-DNA sensor platform previously developed by Dr. Bonham. E-DNA sensors have the potential to serve as affordable, quantitative, and point-of-care tools for clinical diagnosis and academic research, and have been demonstrated to function in crude samples such as whole blood and dirt.
Jody Stephens

Lab Member 2011-2014
Current: Group Lead Software Testing at ArcherDx
Published!
Human transcription factors are proteins that play an important role in the regulation of both normal development and in many diseases. Recent advances in bio-sensors have led to a fluorescent bio-sensor design that uses short sequences of DNA as the sensing element. Ideally, these short sequences fold into two different shapes, with equal numbers of both in solution. These shapes are the same for each of the thousands of different transcription factors. However, the sequence of DNA that generates these shapes is different for each transcription factor. Determining these sequences by hand is unwieldy; therefore, to enable wider use of these bio-sensors an automated way of generating a sequence specific to a transcription factor is necessary. This algorithm needs to efficiently explore the entire search space for candidate solutions while minimizing false negatives and it needs to be able to determine which of these candidate solutions is best. Here, we will demonstrate a partial solution we have implemented using a backtracking tree traversal of the search space and discuss our efforts to refine the algorithm to work on larger sequences and to distinguish between the many candidates it finds. Ultimately, this computational tool will enable the rapid, efficient, and accurate design of bio-sensors that will aid in bio-medical diagnostics and research efforts.
Dr. Ryan Masterson

Lab Member 2012-2013
Current: Resident doctor, Johns Hopkins
Published!
Mason Preusser

Lab Member 2012-2012
Current: Consultant at D-A Lubricant
Cancer and other human diseases are often the result of aberrant gene regulation that arises due to protein-protein interactions – physical associations that may cause conformational change or steric hindrance and play key roles in directing these regulatory processes. Nowhere is this more evident than in the many protein-protein binding events that direct and control the process of transcription, which regulates gene expression in the cell. To better understand the roles of these protein-protein interactions on DNA-binding activity, we are developing sensitive, DNA-based fluorescent biosensors for the quantitative measurement of the functional effects of these interactions. These biosensors allow convenient, quantitative measurement of both the kinetics and thermodynamics of DNA-binding, and may have future roles as medical diagnostic tools for assessing aberrant protein regulation. Here, we use these biosensors to investigate the complex regulation present in the basic helix-loop-helix transcription factors Myc, Max, Sp1, and Miz-1, which associate to form DNA-binding complexes of varying affinity and specificity, and which play key roles in cell growth and cancer progression. Ultimately, these biosensors may allow for earlier and more reliable detection of cancer.
Dr. Tonya Santaus

Lab Member 2012-2012
Current: PhD, University of Maryland Baltimore County
Amanda Faux
Interactions between proteins play key roles in cellular regulation and activity. This is primarily evident in the many protein-protein binding events that direct and control the process of transcription that regulates gene expression in the cell. To better understand the roles of these protein interactions on DNA-binding activity, we are developing sensitive, DNA-based bio-sensors for quantitative measurement of their effects on DNA-protein binding. Specifically, we will be using the a recombinantly expressed and purified form of the basic helix-loop-helix transcription factor Max, which has protein-protein interactions with a variety of protein partners, to investigate changes in DNA binding specificity and affinity.
Matthew Stoddard

Lab Member 2011-2012
Current: Scientific Informatics Consultant at Accenture
Neurogenesis, the process by which neural stem and progenitor cells turn into functional neurons, is heavily regulated. Much of this regulation comes from the interplay of transcription factor proteins that direct the expression of key genes. Two of these transcription factors are the proteins Ascl1 and Lmo4 which regulate genes involved in neuronal commitment and the generation of autonomic neurons among other important genes in development of nervous cells. Ascl1 and Lmo4 mutants are known to be involved in human diseases including human neuroblastomas and small cell lung cancers. Deletion of either gene has resulted in death when tested in mouse models. Many details of the roles Ascl1 and Lmo4 play in neurogenesis, human disease, and how they interact with one another are not fully understood. To address these outstanding questions, we are generating sensitive, DNA-based biosensors for the quantitative detection of the DNA-binding activity of these significant proteins. These sensors will be used with recombinantly expressed and purified constructs of human Ascl1 and Lmo4, with the goal of determining the effect of protein-protein interaction on their regulatory activity.
Morgan Miller

Lab Member 2011-2012
Current: Digital Strategy Director at DMA Digital
Early cancer detection is reliant on sensitive tools to monitor for the presence of abnormal changes in cells. Perhaps one of the most striking features of cancer is the aberrant regulation of transcription factors, proteins that control and direct gene expression. The development of novels tools to assess and understand this transcription factor disfunction is a significant challenge. Here, we present a sensitive, DNA-based bio-sensor that is capable of quantitatively detecting transcription factors involved in cancer onset and progression. These sensors will be used to monitor the DNA-binding activities of the recombinantly expressed and purified proteins Myc, a proto-oncogene up-regulated in many cancers, and Sp1, an enhancer of gene transcription, with the goal of identifying combinatorial effects from the proteins in complex.